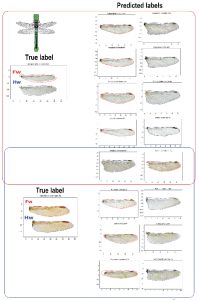
In recent decades, a lack of available knowledge about the magnitude, identity and distribution of biodiversity has given way to a taxonomic impediment where species are not being described as fast as the rate of extinction. Using Machine Learning methods based on seven different algorithms (LR, CART, KNN, GNB, LDA, SVM and RFC) we have created an automatic identification approach for odonate genera, through images of wing contours. The training population is composed of the collected specimens that have been digitized in the framework of the NSF funded Odomatic and TOWD projects. Each contour was pre-processed, and 80 coefficients were extracted for each specimen. These form a database with 4656 rows and 80 columns, which was divided into 70% for training and 30% for testing the classifiers. The classifier with the best performance was a Linear Discriminant Analysis (LDA), which discriminated the highest number of classes (100) with an accuracy value of 0.7337, precision of 0.75, recall of 0.73 and a F1 score of 0.73. Additionally, two main confusion groups are reported, among genera within the suborders of Anisoptera and Zygoptera. These confusion groups suggest a need to include other morphological characters that complement the wing information used for the classification of these groups thereby improving accuracy of classification. Likewise, the findings of this work open the door to the application of machine learning methods for the identification of species in Odonata and in insects more broadly which would potentially reduce the impact of the taxonomic impediment.